Professor Angus LamondFRS FRSE FMedSci
Chair of Biochemistry
Molecular Cell and Developmental Biology, School of Life Sciences

Contact
Research
We are studying gene expression, RNA processing and disease mechanisms in human cells and model organisms. Our goal is to characterise structure/function relationships within the cell and to study protein dynamics at a systems level. We do this using a combination of quantitative techniques, including mass-spectrometry-based proteomics, cell and molecular biology, fluorescence microscopy and computational approaches for big data analytics and interactive data exploration.
The use of high throughput proteomics technologies provides a system-wide approach allowing us to measure cellular responses for a large number of proteins in parallel. An integral part of this work is the development of new software tools for the management analysis and visualisation of complex proteomics data sets.
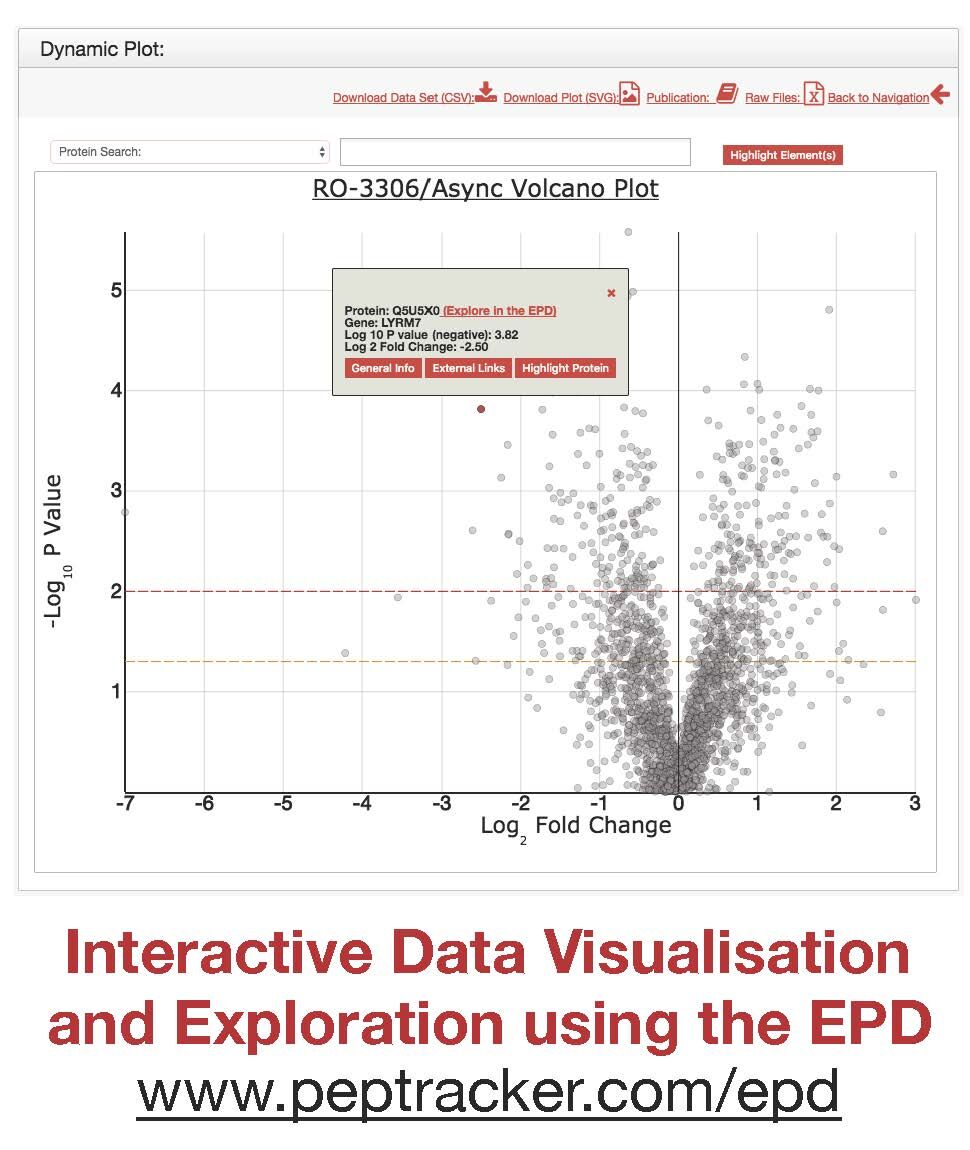
To increase the value of the comprehensive proteomic data sets we have generated, we have incorporated all of our data into an open access, searchable online database, the Encyclopedia of Proteome Dynamics {Brenes:2017vl} (EPD) The EPD provides multiple interactive visualisations, allowing for convenient searching and interactive exploration of all the processed proteomics data. It also provides links to download both the processed data and associated raw MS files, the latter having been deposited in the ProteomeXchange PRIDE repository. The data visualisations in the EPD can be explored by clicking on selected data points, which reveals tooltips providing additional information and links to other related online data resources. Further, using the accompanying Search Box, each data visualization can be searched using multiple selectable criteria, including either proteins/genes of interest, GO terms, subunits of specific protein complexes or relation to functional pathways. Identified components can be readily highlighted and the resulting annotated visualization also downloaded and saved as an editable .svg file. For more detailed descriptions of the functionality and use of the EPD, see Brenes et al., 2017.
In our recent studies we have used combined proteomics, cell biology, microscopy and biochemical approaches in projects including, (i) to characterize the regulation of gene expression during physiological cell cycle progression (ii) to characterize human iPS cells and study genetic variation in the human population, (iii) to study proteome remodeling when healthy human cells are transformed into cancer cells by oncogene activation and (iv) we have used nematodes to study how gene and protein expression responds to acute starvation. In addition, we have studied biochemical mechanisms involved in pre-mRNA processing and alternative RNA splicing and have identified novel small molecule modulators of alternative splicing.
Media availability
I am available for media commentary on my research.
Nuclear structure proteomics and gene expression
Contact Corporate Communications for media enquiries.
Areas of expertise
- Technology
Awards
| Award | Year |
|---|---|
| National Sciences Prizes awarded since 1990 / Appointed British Proteomics Society Lecturer | 2019 |
| Innovation and Knowledge Exchange awards / Scottish EDGE Awards - Platinum Informatics | 2018 |
| Fellow of the Academy of Medical Sciences | 2014 |
| National Sciences Prizes awarded since 1990 / The Novartis Medal & Prize of the Biochemical Society | 2011 |
| Fellow of the Royal Society | 2010 |
| Personal Fellowships / Wellcome Trust Principal Research Fellowship | 2010 |
| Honorary Degrees / dr.scient.h.c. University of Southern Denmark, Syddansk Universitet | 2010 |
| Personal Fellowships / Wellcome Trust Principal Research Fellowship | 2005 |
| Member of the European Molecular Biology Organisation | 1993 |
| National Sciences Prizes awarded since 1990 / The Colworth Medal of the British Biochemical Society | 1992 |
Stories
Press release
Researchers at the University of Dundee are among more than 60 across the UK to receive a share of £12 million to pursue visionary bioscience research.
Press release
Researchers at the University of Dundee have received funding of £4.4 million to explore how our genes can be ‘video-edited’ to develop new drugs for cancer, neurodegeneration and other diseases.
News
The Lamond group have been awarded a £1.128 million BBSRC research grant to support new studies that will dissect RNA processing mechanisms in the nuclei of human cells.