Professor Liz Miller
Chair of Molecular Membrane Biology
Molecular Cell and Developmental Biology, School of Life Sciences

Contact
Biography
Professor Liz Miller joined the Division of Molecular, Cell and Developmental Biology within the School of Life Sciences in 2023. Liz did her PhD in Melbourne, Australia, working on intracellular traffic of plant defence proteins. In 1999 she started a post-doctoral position in Randy Schekman’s lab at the University of California, Berkeley, where she was a Jane Coffin Childs Fellow. In the Schekman lab, Liz established the mechanistic basis for capture of nascent secretory proteins into ER-derived COPII vesicles. In 2005 Liz established her independent lab in the School of Biology at Columbia University in New York. After 10 years at Columbia, Liz moved to become a Programme Leader/MRC Investigator at the MRC Laboratory of Molecular Biology in Cambridge, where she maintains a small lab.
Research
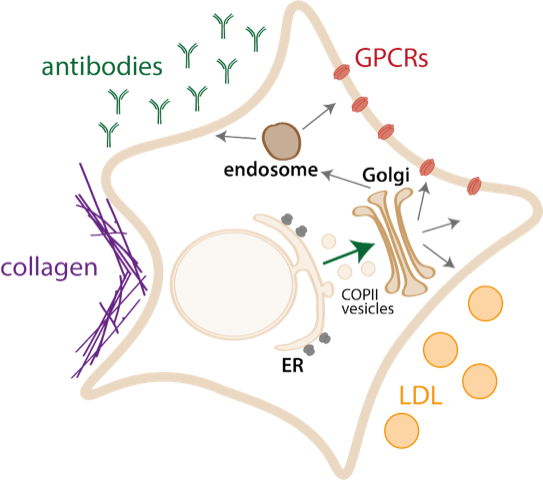
Cellular physiology relies on the accurate deployment of every gene product to the correct cellular compartment. Fully one third of the proteins encoded in eukaryotic genomes navigate the secretory pathway, entering this system via the endoplasmic reticulum (ER). Research in the Miller lab is broadly aimed at understanding basic mechanisms of secretory protein biogenesis, focusing on protein quality control and export from the ER. Historically, we have used the budding yeast, Saccharomyces cerevisiae, as a model system, which affords facile biochemical, genetic, genomic and proteomic tools. By using such a tractable model system, we can rapidly discover new pathways and dissect mechanisms that may be directly relevant to a number of human diseases, most notably cystic fibrosis and similar diseases of protein misfolding. In recent years, we’ve started working with human cell systems, and this will be a major area of development in collaboration with experts within the SLS.
The molecular basis for protein export from the ER via transport vesicles is relatively well understood, and relies on cytoplasmic coat proteins known as the COPII coat. Yet, despite a relatively deep understanding of the mechanisms that drive COPII vesicle formation, we still don’t understand the molecular interactions that drive specific cargo capture, and how this process is regulated to prevent improper traffic of misfolded proteins. We aim to solve this mechanistic problem of an ER export code, where transmembrane receptors bind to cargo proteins and in turn interact directly with the COPII coat to drive export. By understanding the molecular details of these protein-protein interactions, we hope to develop small molecules that selectively inhibit secretion of some proteins but leave the rest of the secretome in tact.
People in my lab
- Xiaohan Li (Dundee)
- Jasmine Pham (Dundee)
- Laura Spinelli (Dundee)
- Julijia Maldutyte (Cambridge)
- Sarah Triclin (Cambridge)
- Ying Bai (Cambridge)
- Ivan Domeneche Mercade (Cambridge)
Selected Publications
- Melero, A., Boulanger, J., Kukulski, W. and Miller, E.A. (2022) Ultrastructure of COPII vesicle formation in yeast characterized by correlative light and electron microscopy. Mol. Biol. Cell. doi:10.1091/mbc.E22-03-0103
- Gomez-Navarro, N., Maldutyte, J., Poljak, K., Peak-Chew, S-Y., Orme, J., Bisnett, B.J., Lamb, C.H., Boyce, M., Gianni, D. and Miller, E.A. (2022) Selective inhibition of protein secretion by abrogating receptor-coat interactions during ER export. Proc. Natl. Acad. Sci, USA. 119(31):e2022080119 doi:10.1073/pnas.2202080119
- Stancheva, V.G., Li, X-H., Hutchings, J., Gomez-Navarro, N., Santhanam, B., Madan Babu, M., Zanetti, G. and Miller, E.A. (2020) Combinatorial multivalent interactions drive cooperative assembly of the COPII coat. J. Cell Biol. 219(11):e202007135
- Gomez-Navarro, N., Melero, A., Li, X-H., Boulanger, J., Kukulski, W. and Miller, E.A. (2020) Cargo crowding contributes to sorting stringency in COPII vesicles. J. Cell. Biol. 219(7):e201806038
- Lakshminarayan, R., Phillips, B.P., Binnian, I.L., Gomez-Navarro, N., Erscudero-Urquijo, N., Warren, A.J. and Miller, E.A. (2020) Pre-emptive quality control of a misfolded membrane protein by ribosome-driven effects. Current Biology 30(5):854-864
- D’Arcangelo, J.G., Crissman, J., Pagant, S., Copic, A., Latham, C. Snapp, E.L. and Miller, E.A. (2015) Traffic of p24 Proteins and COPII Coat Composition Mutually Influence Membrane Scaffolding. Current Biology, 25(18):1296-1305.
- Pagant, S., Wu, A., Edwards, S. and Miller, E.A. (2015) Sec24 is a coincidence detector that binds both cargo proteins and the cargo receptor, Erv14, to drive ER export. Current Biology 25(4):403-412.
- Louie, R.J., Guo, J., Rodgers, J.W., White, R., Shah, N., Pagant, S., Kim, P., Livstone, M., Dolinski, K., McKinney, B., Hong, J., Sorscher, E.J., Bryan, J., Miller, E.A. and Hartman, J.L. (2012) A yeast phenomic model for the gene interaction network modulation CFTR-DF508 protein biogenesis. Genome Med. 4(12):103
- Copic, A., Latham, C.F., Horlbeck, M., D’Arcangelo, J.G. and Miller, E.A. (2012) ER cargo properties specify a requirement for COPII coat rigidity mediated by Sec13p. Science 335(6074):1359-1362.
Awards
| Award | Year |
|---|---|
| Major Personal Funding Awards / Wellcome Discovery Award | 2022 |
Stories
News
Professor Liz Miller from the School of Life Sciences has been awarded funding from the Medical Research Council to study how cells regulate protein secretion. Secreted proteins perform diverse functions important for human health.