
Contact
Websites
Biography
I studied cyanobacteria for my PhD at the University of Dundee with Pete Rowell and Sir William Stewart. I first worked on RNA with John Brown in collaboration with Jean Beggs from the University of Edinburgh. I won a Royal Society fellowship to join Witold Filipowicz’s lab at the Friedrich Miescher Institute, Basel, Switzerland and later joined Dame Caroline Dean’s lab at the John Innes Centre. I returned to the University of Dundee as a Lecturer, Reader, and then Professor of Molecular Genetics.
Research
RNA processing and modification
We study RNA. Although the genes in our chromosomes are made of DNA, they are transcribed into copies of RNA when switched on. RNA is processed and modified in ways that ultimately determine what a gene codes for. We study processing events called splicing, polyadenylation, and RNA modification. In this way, we learn what genomes encode and how the regulation of genes leads to changes in biology.
Our science is curiosity-led and attempts to answer basic questions about RNA and biology. We try to use unbiased approaches that include forward genetic screens, global sequencing of messenger RNAs and inter-species association mapping. We combine expertise in RNA molecular biology with genetics and computational biology.
Genetic screens to understand flower development led us to study RNA binding proteins that affect splicing, polyadenylation and the methylation of RNA. Although this work started with Arabidopsis, we switch experimental systems to generalise our understanding of the most fundamental questions we can. Since sequencing RNAs reveals what genomes encode, we have partnered with the African Orphan Crop Consortium to directly sequence RNA from indigenous crops, including yam, to provide a resource to help develop future food security.
We try to be as transparent as possible in our scientific process. We freely share all our source data and the code we use to analyse our data sets. We pre-print all our work and publish it in open-access scientific journals with transparent peer review so anyone can read it for free. Our public engagement activities include a genetics garden we established at the University of Dundee Botanic Garden more than 10 years ago.
Our work is publicly funded by the UK government through BBSRC and GCRF, and by the EU through Horizon 2020.
Selected Publications
Shen, A., Hencel, K., Parker, M.T., Scott, R.,Skukan, R., Adesina, A.S., Metheringham, C.L., Miska, E.A., Nam, Y., Haerty, W., Simpson, G.G., Akay, A. (2024) U6 snRNA m6A modification is required for accurate and efficient splicing of C. elegans and human pre-mRNAs. Nucleic Acids Research 2024 gkae 447 https://doi.org/10.1093/nar/gkae447
Parker, M. T., Fica, S. M., Barton, G. J. & Simpson, G. G. (2023) Inter-species association mapping links splice site evolution to METTL16 and SNRNP27K. eLife, 12, e91997. https://elifesciences.org/articles/91997
Parker, M. T., Soanes, B. K., Kusakina, J., Larrieu, A., Knop, K., Joy, N., Breidenbach, F., Sherwood, A. V., Barton, G. J., Fica, S. M., Davies, B. H., & Simpson, G. G. (2022). m6A modification of U6 snRNA modulates usage of two major classes of pre-mRNA 5’ splice site. eLife, 11, e78808. https://elifesciences.org/articles/78808
Zhang, M., Bodi, Z., Mackinnon, K., Zhong, S., Archer, N., Mongan, N. P., Simpson, G. G., & Fray, R. G. (2022). Two zinc finger proteins with functions in m6A writing interact with HAKAI. Nature Communications, 13(1), 1127. https://doi.org/10.1038/s41467-022-28753-3
Bredeson, J.V., Lyons, J.B., Oniyinde, I.O., Okereke, N.R., Kolade, 0., Nnabue, I., Nwadili, C.O., Hřibová,E., parker, M.T.,Nwogha,J., Shu, S., Carlson, J., Kariba, R., Muthemba, S., knop, K., Barton, G.J., Sherwood, A.V., Lopez-Montes, A., Asiedu, R., Jamnadass, R., Muchugi, A., Goodstein, D., Egesi, C.N., Featherston, J., Asfaw, A., Simpson, G.G., Doležel, J., Hendre, P.S., Van Deynze, A., Kumar, P.L., Obidiegwu, J.E., Bhattacharjee, R., Rokhsar, D.S. (2022). Chromosome evolution and the genetic basis of agronomically important traits in greater yam. Nature Communications, 13(1), 2001. https://doi.org/10.1038/s41467-022-29114-w
Parker, M. T., Barton, G. J., & Simpson, G. G. (2021). Yanocomp: robust prediction of m6A modifications in individual nanopore direct RNA reads. In bioRxiv (p. 2021.06.15.448494). https://doi.org/10.1101/2021.06.15.448494
Parker, M. T., Knop, K., Zacharaki, V., Sherwood, A. V., Tomé, D., Yu, X., Martin, P. G., Beynon, J., Michaels, S. D., Barton, G. J., & Simpson, G. G. (2021). Widespread premature transcription termination of Arabidopsis thaliana NLR genes by the spen protein FPA. eLife, 10, e65537. https://doi.org/10.7554/eLife.65537
Parker, M. T., Knop, K., Barton, G. J., & Simpson, G. G. (2021). 2passtools: two-pass alignment using machine-learning-filtered splice junctions increases the accuracy of intron detection in long-read RNA sequencing. In Genome Biology 22, 72. https://doi.org/10.1186/s13059-021-02296-0
Parker, M. T., Knop, K., Sherwood, A. V., Schurch, N. J., Mackinnon, K., Gould, P. D., Hall, A. J., Barton, G. J., & Simpson, G. G. (2020). Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. eLife, 9, e49658. https://doi.org/10.7554/eLife.49658
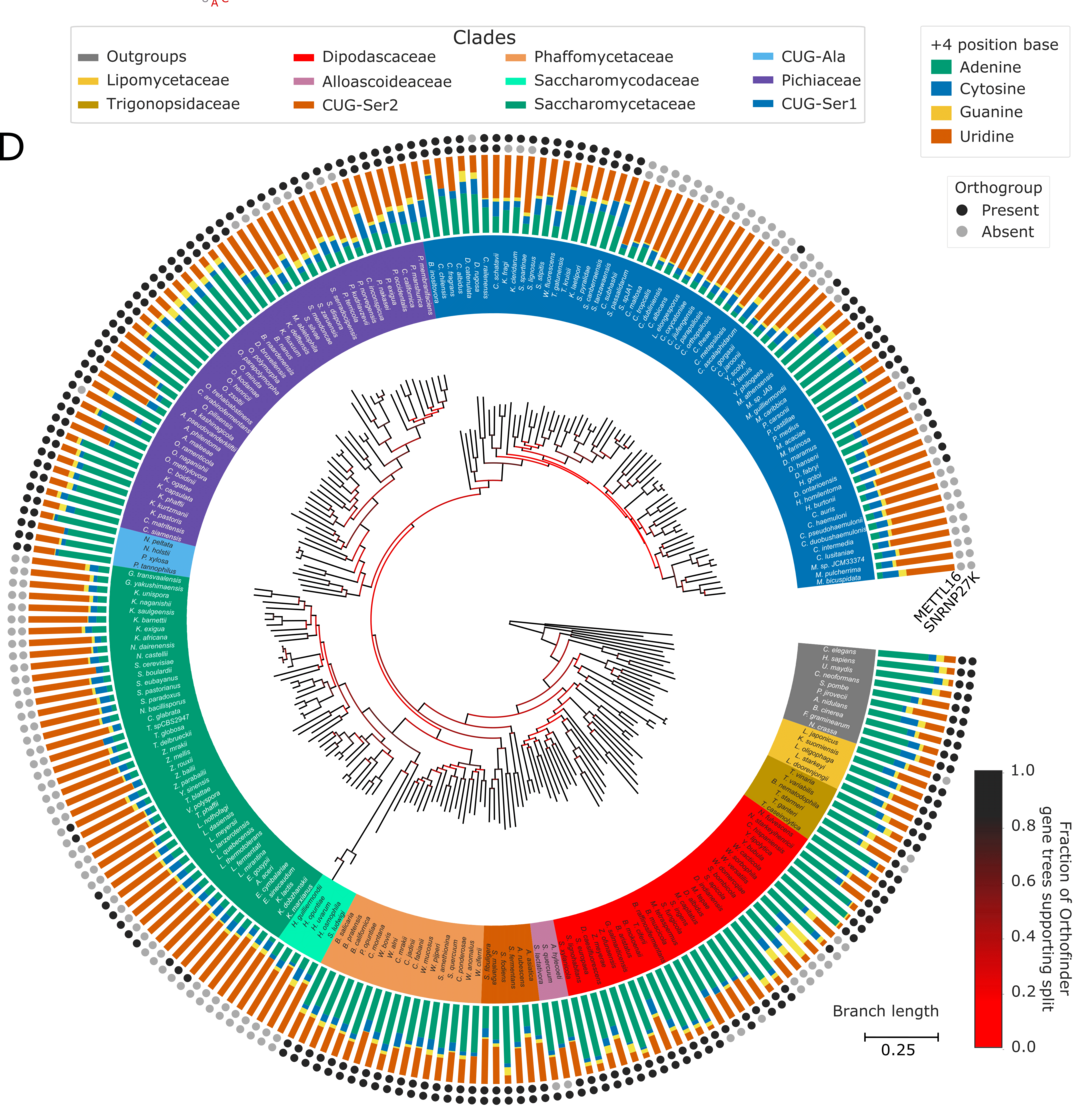
Figure 1: We used inter-species association mapping to show that the evolution of 5’ splice site sequence preference is linked to the splicing factor SNRNP27K and the enzyme METTL16, which methylates U6snRNA. From Parker et al., 2023.
Teaching
I teach Genetics and Advanced Plant Science. My lab hosts final year Honours students carrying out their research projects, and Masters and Erasmus students from overseas.
Awards
| Award | Year |
|---|---|
| Fellow of the Royal Society of Biology | 2015 |
Stories

News
New work from the University of Dundee reveals a basic feature of how our genomes are organized, and our genes are processed. This work has been published in the journal eLife.
News
Researchers in the School of Life Sciences at the University of Dundee have helped to uncover and understand the genome of a vitally important orphan crop, called water yam.
News
A study published this week in the journal eLife, by a team at University of Dundee's School of Life Sciences uses a new approach to reveal the complexity and modifications of RNA that are essential to genetic control.