Dr Fede Pelisch
Principal Investigator/Senior Lecturer
Molecular Cell and Developmental Biology, School of Life Sciences

Contact
Biography
Fede Pelisch is Principal Investigator within the Division of Molecular Cell & Developmental Biology. He obtained a PhD in from the University of Buenos Aires, Argentina, in 2010. He worked as a post-doctoral researcher, first at the University of Buenos Aires, and later, from 2012, he was awarded a Marie Curie International Fellowship to join the University of Dundee. During this time, Fede received the Molecular and Cellular Biology Prize (School of Life Sciences) and was later awarded an MRC Career Development Award to start his own research group.
Research
My lab started in 2018 funded by a Career Development Fellowship from the Medical Research Council. My group is interested in understanding the mechanisms regulating chromosome segregation during meiosis and mitosis. We use the nematode Caenorhabditis elegans as a model system (Figure 1) and we employ a variety of methodologies including biochemical analysis, high-resolution imaging, and a range of genetics tools. Since then, we focused on understanding the role of dynamic phosphorylation and dephosphorylation during chromosome segregation in oocyte meiosis.
By combining high-resolution microscopy with biochemical methods, we described different roles for the phosphatase PP2A/B56 during oocyte meiosis and identified the mechanism of PP2A/B56 targeting to chromosomes in specific meiotic stages (Bel Borja et al. 2020).
We used chemical genetics to describe the different roles of polo-like kinase 1 (PLK1) during oocyte meiosis while at the same time characterised its targeting mechanism to meiotic chromosomes (Taylor et al. 2023). We are currently following up on these findings focusing on the identification of key substrates for both PLK-1 and PP2A/B56.
In our latest work, we characterised an inner kinetochore binding site for PLK1 and its relevance during the first mitotic embryonic division. We found that PLK1 targeted by the inner kinetochore protein CENP-C regulates chromosome congression and mitotic timing without regulating the spindle assembly checkpoint (Bel Borja et al. 2024). These findings provide the foundation for better understanding how different populations of PLK1 perform distinct functions during mitosis.
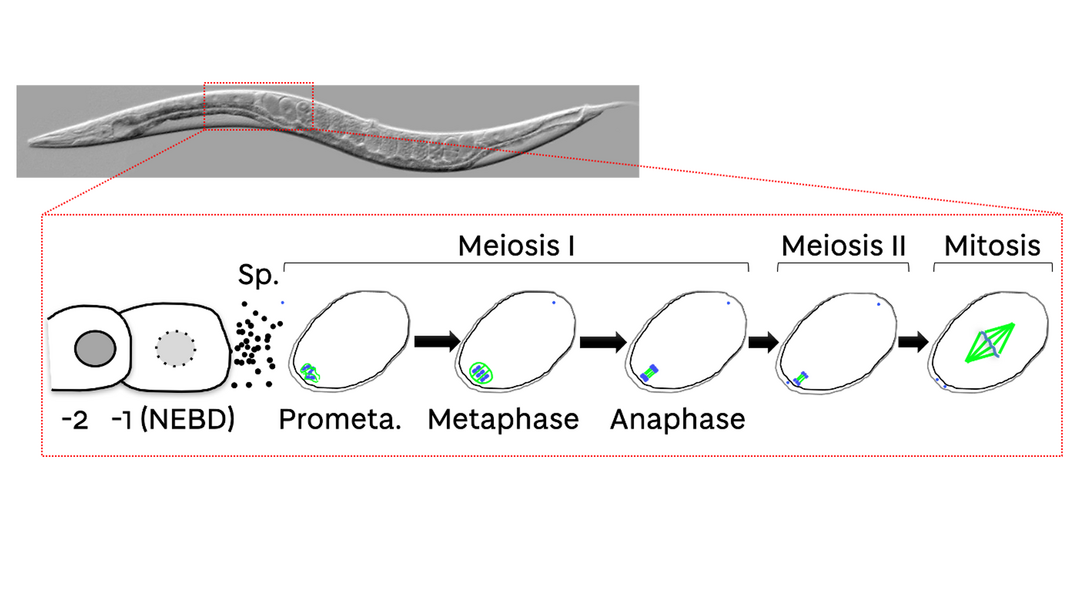
Figure 1: C. elegans oocytes and embryos. A representative image of a C. elegans worm is shown. The schematic below highlights the region of the worm where the meiotic and first mitotic divisions occur.
Selected Publications
- Bel Borja, L.; Taylor, S.J.P.; Soubigou, and Pelisch, F*. CENP-C-targeted PLK-1 regulates kinetochore function in C. elegans embryos. bioRxiv (2024) 04.26.591339; doi: https://doi.org/10.1101/2024.04.26.591339
- Taylor, S.J.P.; Bel Borja, L.; Soubigou, F.; Houston, J.; Cheerambathur, D.K.; and Pelisch, F*. BUB-1 and CENP-C recruit PLK-1 to Control Chromosome Alignment and Segregation During Meiosis I in C. elegans Oocytes. eLife 12:e84057 (2023) doi: 10.7554/eLife.84057
- Bel Borja, L.; Soubigou, F.; Taylor, S.J.P.; Fraguas Bringas, C.; Budrewicz, J.; Lara-Gonzalez, P.; Sorensen Turpin, C.G.; Bembenek, J.N.; Cheerambathur, D.K.; and Pelisch, F*. BUB-1 targets PP2A:B56 to regulate chromosome congression during meiosis I in C. elegans oocytes. eLife 9:e65307 (2021) doi: 10.7554/eLife.65307.
- Taylor, S.J.P and Pelisch, F*. Chromosome segregation during female meiosis in C. elegans: a tale of pushing and pulling. Journal of Cell Biology (2020); doi: 10.1083/jcb.202011035.
- Pelisch, F.* et al. Sumoylation regulates protein dynamics during meiotic chromosome segregation in oocytes. Journal of Cell Science, 2019. 132(14): p.jcs232330. doi: 10.1242/jcs.232330
- Pelisch, F. et al. A SUMO-dependent protein network regulates chromosome congression during oocyte meiosis. Molecular Cell (2017), 65:66-77. doi: 10.1016/j.molcel.2016.11.001
- Pelisch, F.* & Hay, R.T. Tools to study SUMO conjugation in C. elegans. Methods Mol Biol. 2016;1475:233-56. doi: 10.1007/978-1-4939-6358-4_17.
- Pelisch, F. et al. Dynamic SUMO modification regulates mitotic chromosome assembly and cell cycle progression in Caenorhabditis elegans. Nature Communications (2014) Dec 5;5:5485. doi: 10.1038/ncomms6485.
Public Engagement
I am leading the new partnership of the MCDB division with the Harris Academy. This partnership will provide Advanced Higher Biology students the opportunity to visit the Life Sciences building and see how it operates, from support staff to research scientists. Students receive a lecture on topics related to their curriculum and visit the state-of-the-art facilities in the School.
PhD Projects
Principal supervisor
PhD opportunity
Awards
| Award | Year |
|---|---|
| Personal Fellowships / MRC Career Development Fellowship | 2017 |
| Molecular and Cellular Biology Prize | 2016 |
Stories
News
Federico (Fede) Pelisch has been appointed to the eLife Board of Directors.